Pynta¶
pynta is an automated workflow code to enable the calculation of thermochemistry
and rate coefficients for reactions involving metallic surfaces.
The pynta code is designed to automatically characterize chemical reactions
relevant to heterogeneous catalysis. In particular, it spawns and processes a
large number of ab initio quantum chemistry calculations to study gas-phase
reactions on crystal facets. It is designed to run both on local clusters and
on petascale and upcoming exascale machines.
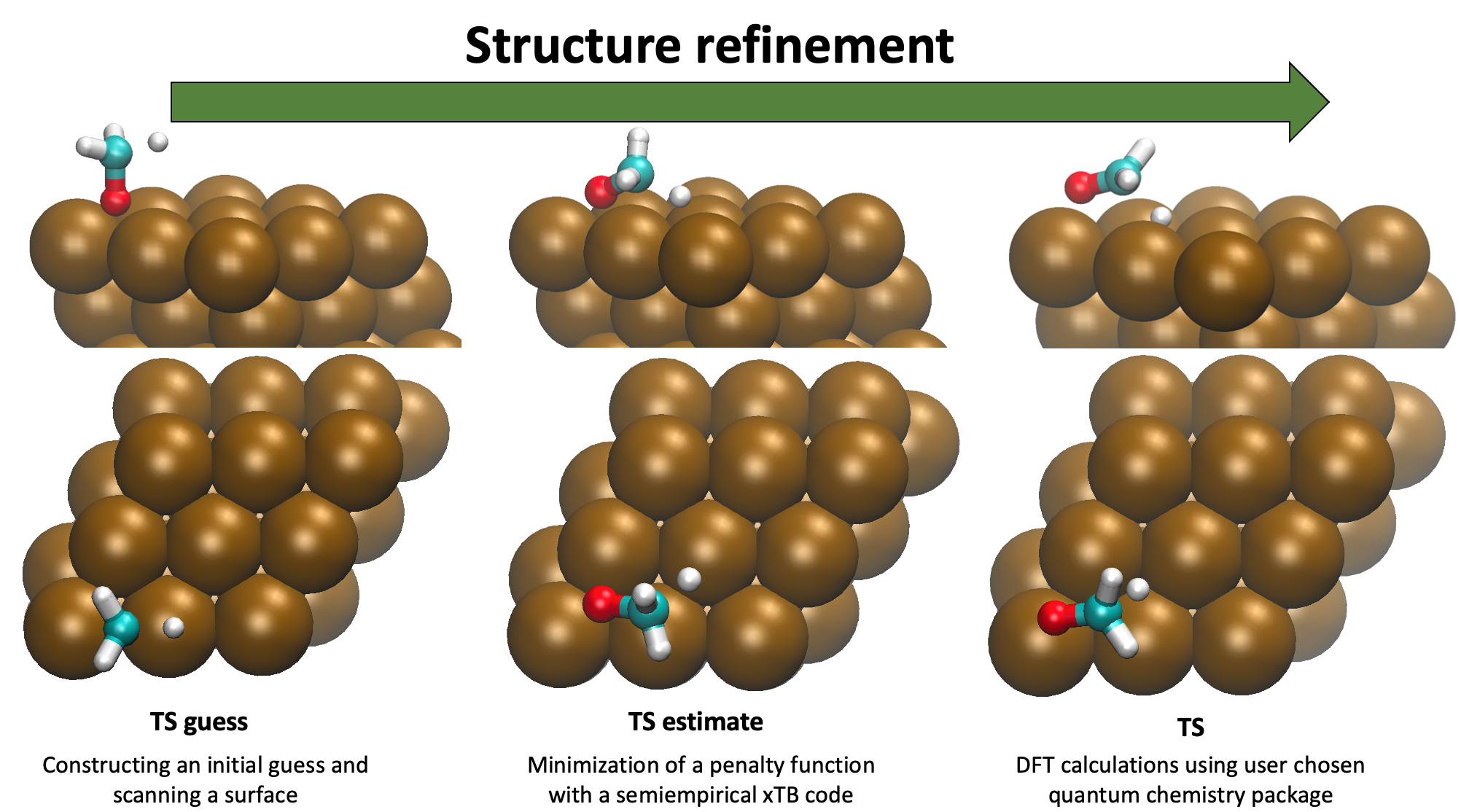
Pynta generates initial guesses for all gas phase species and adsorbates on the surface accounting for the symmetry of the surface. These guesses are then optimized and frequencies are calculated for the unique resulting geometries. Based on the provided reaction semi-atom mappings the adsorbate are placed on the surface and perturbed in many unique ways to generate transition state guesses, the best guesses are optimized and vibrational frequencies and IRCs are generated for unique successfully optimized transition states. This information can then be processed to generate thermochemistry and rate coefficients for the adsorbates and reactions that can be used in microkinetic models.
Pynta is designed to work with the workflow code Fireworks , which provides users with significant flexiblity in running Pynta.
Installation¶
Notes on Installation¶
Pynta supports only Linux and MacOS installation and currently only provides installation from source.
Install git¶
If you do not already have git it needs to be installed. On linux install with the appropriate package manager. On MacOS git should be installed through XCode Commandline Tools. This can be done by simply running git if it is not installed it will prompt you to install XCode Commandline Tools.
Install Anaconda¶
Download the appropriate Anaconda or Miniconda . For the command line/bash installers find the downloaded .sh file and run it with bash for example on linux this might look like:
bash Anaconda3-2022.10-Linux-x86_64.sh
Make sure to tell the installer to append Anaconda/Miniconda to PATH when asked.
After installation you will need to exit and reopen your command line interface. After doing so we (optionally) recommend that you install mamba to speed up a later part of the install:
conda install mamba
Install the Pynta source code:¶
git clone git@github.com:zadorlab/pynta.git
Create a conda environment for Pynta¶
Enter the Pynta source directory with:
cd pynta
and then create the conda environment. If you have installed mamba run
mamba env create -f environment.yml
else you can instead run the slower (but equally valid) commmand:
conda env create -f environment.yml
Once this is done activate the generated environment with:
conda activate pynta_env
Install MongoDB¶
The code for fireworks itself is installed within the created pynta_env, however Fireworks requires a MongoDB instance to manage workflows. There are many different ways to configure this and the best way depends heavily on where and how you are running Pynta. The community edition of MongoDB can be installed on a server or local computer you own, directions are available here . Additionally a MongoDB can be setup with a cloud provider. MongoDB provides a free option for their MongoDB Atlas that should be sufficient for typical use of Fireworks.
Setup Fireworks¶
Once the MongoDB is setup we can properly configure Fireworks. There are four configuration files that need written for Fireworks: my_launchpad.yaml,my_fworker.yaml, my_qadapter.yaml and FW_config.yaml. The first, my_launchpad.yaml needs setup for your particular MongoDB, an example is provided in the last section of this page of the fireworks documentation. The second, my_fworker.yaml can typically just be:
name: my first fireworker
category: ''
query: '{}'
The third my_qadapter.yaml is not necessary if you don’t intend to have Fireworks submit jobs through a queue. A description of how this operates within Fireworks is available in their documentation here . Examples of my_qadapter.yaml are available here . Lastly the FW_config.yaml file should be configured according to directions in the Fireworks documentation here . After configuring fireworks you should be able (after activating the pynta_env) run:
lpad get_wflows
Fireworks has a lot of handy features that enable you to track workflow progress and statistics in their gui and commands that enable you to manually launch and pause components of the workflow. Their documentation is available here .
Testing Pynta¶
Pynta has a set of unit and functional tests that can be run to ensure Pynta is properly installed and executes properly. In the Pynta directory all tests can be run with
make test-all
only unittests can be run with
make test-unitests
and only functional tests can be run with
make test-functional
Warning one of the functional tests runs a small, but structurally complete Pynta workflow. Depending on the speed of the computer this can take a half hour to several hours to run.
Running Pynta¶
In order to run Pynta we first need to describe the reactions we want Pynta to calculate. We do this within a reactions.yaml file. This file describes the reactants, products and the atom mapping between them in the RMG adjacency list format . All atoms that are part of bonds that break and form in the reaction need to be labeled (with the same label) in both reactants and products. The validity of adjacency lists can be checked using RMG’s tool here .
Example reactions.yaml file¶
- index: 0
reactant: 'multiplicity 1
1 *3 O u0 p2 c0 {2,S} {5,S}
2 *2 C u0 p0 c0 {1,S} {3,S} {4,S} {7,S}
3 H u0 p0 c0 {2,S}
4 H u0 p0 c0 {2,S}
5 *4 H u0 p0 c0 {1,S}
6 *1 X u0 p0 c0
7 *5 X u0 p0 c0 {2,S}
'
product: 'multiplicity 1
1 *3 O u0 p2 c0 {2,S} {6,S}
2 *2 C u0 p0 c0 {1,S} {3,S} {4,S} {5,S}
3 H u0 p0 c0 {2,S}
4 H u0 p0 c0 {2,S}
5 *4 H u0 p0 c0 {2,S}
6 *1 X u0 p0 c0 {1,S}
7 *5 X u0 p0 c0
'
reaction: OC[Pt] <=> CO[Pt]
reaction_family: Surface_Migration
- index: 1
reactant: 'multiplicity 1
1 *3 X u0 p0 c0 {2,D}
2 *1 O u0 p2 c0 {1,D}
3 *2 H u0 p0 c0 {4,S}
4 *4 X u0 p0 c0 {3,S}
'
product: 'multiplicity 1
1 *3 X u0 p0 c0 {2,S}
2 *1 O u0 p2 c0 {1,S} {3,S}
3 *2 H u0 p0 c0 {2,S}
4 *4 X u0 p0 c0
'
reaction: '[Pt] + O[Pt] <=> O=[Pt] + [H][Pt]'
reaction_family: Surface_Dissociation
Calling Pynta¶
An example python script for calling Pynta is available below.
The Pynta function has many parameters. It is best to divide them based on what they are associated with. First there are the run parameters:
path: the directory in which Pynta will execute and save files
rxns_file: the location of the reactions.yaml file containing the reactions to calculate
Second there are the Fireworks parameters:
launchpad_path: the path to the my_launchpad.yaml file, by default it will use your default my_launchpad.yaml
fworker_path: the path to the my_fworker.yaml file, by default it will use your default my_fworker.yaml
queue_adapter_path: the path to the my_qadapter.yaml file, by default it will use your default my_qadapter.yaml
reset_launchpad: if true Pynta will reset the Fireworks launchpad during construction, this will delete any existing workflows on fireworks and is usually undesirable
num_jobs: the number of jobs for Pynta to launch if running on multiple nodes outside a queue
queue: if true will run Fireworks in queue mode using the file at queue_adapter_path
Third there are the slab specification parameters:
metal: the identity of the slab metal ex: Cu, Pt
surface_type: the facet ex: fcc111, bcc110
vacuum: the height of the vacuum in Angstroms above the slab in the cell
repeats: in general operation this should be [(1,1,1),(x,y,z)] where x,y,z are the number of atoms in the x,y and z directions in the slab
a: the lattice constant of the metal, if not specified and slab_path is not specified Pynta will calculate it
slab_path: path to the geometry of the slab, this will cause Pynta to skip slab generation, but must be consistent with metal, surface_type, vacuum and repeats
Fourth there are the ASE parameters that control the quantum chemistry calculation execution. All of these parameters need to be specified in terms of what ASE expects.
software: this is the string corresponding to an ASE calculator object. Pynta will search ASE for a calculator with this name.
software_kwargs: this is the dictionary of keyword arguments passed to the ASE calculator object on construction. This particular dictionary is the default set of arguments.
software_kwargs_gas: this is a dictionary of keyword arguments that should be different from software_kwargs when running gas phase calculations
TS_opt_software_kwargs: this is a dictionary of keyword arguments that should be different from software_kwargs when running saddle point optimizations
lattice_opt_software_kwargs: this is a dictionary of keyword arguments that should be different from software_kwargs when optimizing the lattice constant
Lastly there are the Pynta energy filter criteria:
Eharmtol: a tolerance such that all TS/adsorbate guesses are always calculated if they have energies less than Emin * Eharmtol
Eharmfiltertol: a tolerance such that all TS/adsorbate guesses are never calculated if they have energies greater than Emin * Eharmfiltertol
Ntsmin: the minimum number of TS/adsorbate guesses. If the number of guesses with energies less than Emin * Eharmtol is less than Ntsmin Pynta will add the lowest energy guesses (that are less than Emin * Eharmfiltertol) until it has Ntsmin guesses.
Pynta’s execute function has a few important options:
generate_initial_ad_guesses: if False Pynta will not generate initial adsorbate guesses and assume they are already in the appropriate directories.
calculate_adsorbates: if False Pynta will not generate Fireworks jobs for calculating adsorbates and will assume the results are already in the appropriate directories.
calculate_transition_states: if False Pynta will not generate Fireworks jobs for finding and calculating transition states
launch: if True Pynta will attempt to launch the Fireworks workflow in infinite mode after it is constructed. If False the workflow is still generated and added to the launchpad, but it is left up to the user to handle launching through Fireworks commands.
Modules¶
- class pynta.calculator.HarmonicallyForcedXTB(atoms: Optional[Atoms] = None, **kwargs)[source]¶
Bases:
XTB
- pynta.calculator.add_sella_constraint(cons, d)[source]¶
construct a constraint from a dictionary that is the input to the constraint constructor plus an additional “type” key that indices the name of the constraint in this case for Sella the full Constraints object cons must be included in the inputs adds the constraint to Constraints and returns None
- pynta.calculator.get_lattice_parameter(metal, surface_type, software, software_kwargs, da=0.1, options={'xatol': 0.0001})[source]¶
- pynta.calculator.run_harmonically_forced_xtb(atoms, atom_bond_potentials, site_bond_potentials, nslab, method='GFN1-xTB', constraints=[])[source]¶
Optimize TS guess using xTB + harmonic forcing terms determined by atom_bond_potentials and site_bond_potentials
- class pynta.main.Pynta(path, rxns_file, surface_type, metal, label, launchpad_path=None, fworker_path=None, vacuum=8.0, repeats=[(1, 1, 1), (3, 3, 4)], slab_path=None, software='Espresso', socket=False, queue=False, njobs_queue=0, a=None, software_kwargs={'conv_thr': 1e-06, 'degauss': 0.01, 'ecutwfc': 40, 'kpts': (3, 3, 1), 'mixing_mode': 'local-TF', 'nosym': True, 'occupations': 'smearing', 'pseudopotentials': {'C': 'C.pbe-n-kjpaw_psl.1.0.0.UPF', 'Cu': 'Cu.pbe-spn-kjpaw_psl.1.0.0.UPF', 'H': 'H.pbe-kjpaw_psl.1.0.0.UPF', 'N': 'N.pbe-n-kjpaw_psl.1.0.0.UPF', 'O': 'O.pbe-n-kjpaw_psl.1.0.0.UPF'}, 'smearing': 'marzari-vanderbilt', 'tprnfor': True}, software_kwargs_gas=None, TS_opt_software_kwargs=None, lattice_opt_software_kwargs={'degauss': 0.02, 'ecutwfc': 70, 'kpts': (25, 25, 25), 'mixing_mode': 'plain'}, reset_launchpad=False, queue_adapter_path=None, num_jobs=25, max_num_hfsp_opts=None, Eharmtol=3.0, Eharmfiltertol=30.0, Ntsmin=5)[source]¶
Bases:
object- execute(generate_initial_ad_guesses=True, calculate_adsorbates=True, calculate_transition_states=True, launch=True)[source]¶
generate and launch a Pynta Fireworks Workflow if generate_initial_ad_guesses is true generates initial guesses, otherwise assumes they are already there if calculate_adsorbates is true generates firework jobs for adsorbates, otherwise assumes they are not needed if calculate_transition_states is true generates fireworks jobs for transition states, otherwise assumes they are not needed if launch is true launches the fireworks workflow in infinite mode…this generates a process that will continue to spawn jobs if launch is false the Fireworks workflow is added to the launchpad, where it can be launched separately using fireworks commands
- generate_initial_adsorbate_guesses(skip_structs=False)[source]¶
Generates initial guess geometries for adsorbates and gas phase species Generates maps connecting the molecule objects with these adsorbates
- generate_mol_dict()[source]¶
generates all unique Molecule objects based on the reactions and generates a dictionary mapping smiles to each unique Molecule object also updates self.rxns_dict with addtional useful information for each reaction
- generate_slab()[source]¶
generates and optimizes a small scale slab that can be scaled to a large slab as needed optimization occurs through fireworks and this process waits until the optimization is completed
- setup_adsorbates(initial_guess_finished=False)[source]¶
Attaches each adsorbate structure to the slab and sets up fireworks to first optimize each possible geometry and then run vibrational calculations on each unique final geometry
- setup_transition_states(adsorbates_finished=False)[source]¶
Sets up fireworks to generate and filter a set of TS estimates, optimize each unique TS estimate, and run vibrational and IRC calculations on the each unique final transition state Note the vibrational and IRC calculations are launched at the same time
- pynta.mol.add_adsorbate_to_site(atoms, adsorbate, surf_ind, site, height=None, orientation=None, tilt_angle=0.0)[source]¶
The base function for adding one adsorbate to a site. Site must include information of ‘normal’ and ‘position’. Useful for adding adsorbate to multiple sites or adding multidentate adsorbates.
- Parameters:
atoms (ase.Atoms object) – Accept any ase.Atoms object. No need to be built-in.
adsorbate (str or ase.Atom object or ase.Atoms object) – The adsorbate species to be added onto the surface.
site (dict) – The site that the adsorbate should be added to. Must contain information of the position and the normal vector of the site.
height (float, default None) – The height of the added adsorbate from the surface. Use the default settings if not specified.
orientation (list or numpy.array, default None) – The vector that the multidentate adsorbate is aligned to.
tilt_angle (float, default None) – Tilt the adsorbate with an angle (in degrees) relative to the surface normal.
- pynta.mol.generate_adsorbate_guesses(mol, ads, slab, repeats, cas, mol_to_atoms_map, metal, single_site_bond_params_lists, single_sites_lists, double_site_bond_params_lists, double_sites_lists, Eharmtol, Eharmfiltertol, Ntsmin)[source]¶
- pynta.mol.generate_unique_site_additions(geo, cas, nslab, site_bond_params_list=[], sites_list=[])[source]¶
- pynta.mol.get_ase_index(ind, template_mol_map, molecule_to_gratom_maps, nslab, ads_sizes)[source]¶
get the index associated with the ase.Atoms object
- pynta.mol.get_bond_lengths_sites(mol, ads, atom_map, surf_atom_map, nslab, facet='fcc111', metal='Cu', cas=None)[source]¶
gets bond lengths and site information indexed to the Molecule object atoms bond lengths is a matrix with the distances between all atoms that share bonds site information is the atom index mapped to the site type
- pynta.mol.get_broken_formed_bonds(reactant, product)[source]¶
compares the reactant and product structures assuming bonds only form and break between labeled atoms returns frozensets of pairs of labels associated with atoms whose bond breaks or is formed during the reaction
- pynta.mol.get_edges(slab_path, find_surface=False)[source]¶
Get adsorption edges
- Parameters:
find_surface (bool) – specify whether to include surface or not default = False
- Returns:
edges (list[tuple]) – adsobrtion edges
surface (numpy.ndarray) – an array with tags describing: top surface atoms (1) bottom surface (-1) and bulk atoms (0) Atoms with tags ‘1’ are considered as the possible binding spots
- pynta.mol.get_labeled_bonds(mol)[source]¶
generate a list of all Bonds between labeled atoms in the Molecule object
- pynta.mol.get_mol_index(ind, template_mol_map)[source]¶
ind is the index in the template first index corresponds to the molecule object second index corresponds to the index in the molecule object
- pynta.mol.get_nonintersectingkeys_maps(maps)[source]¶
process the subgraph isomorphisms found to generate a list of valid maps
- pynta.mol.get_template_mol_map(template, mols)[source]¶
template is the combined labeled Molecule object find dictionary mapping indices of the template to the molecule objects of interest output is a list with each item coresponding to a Molecule object in mols the dictionary maps indices of the template to indices of the corresponding mol object
- class pynta.tasks.CollectTask(*args, **kwargs)[source]¶
Bases:
FiretaskBase- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.DoNothingTask(*args, **kwargs)[source]¶
Bases:
FiretaskBase- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.EnergyTask(*args, **kwargs)[source]¶
Bases:
FiretaskBase- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- pynta.tasks.IRC_firework(xyz, label, out_path=None, spawn_jobs=False, software=None, socket=False, software_kwargs={}, opt_kwargs={}, run_kwargs={}, constraints=[], parents=[], ignore_errors=False, forward=True)[source]¶
- class pynta.tasks.MolecularCollect(*args, **kwargs)[source]¶
Bases:
CollectTask- required_params = ['xyzs', 'check_symm', 'fw_generators', 'fw_generator_dicts', 'out_names', 'future_check_symms', 'label']¶
- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.MolecularEnergyTask(*args, **kwargs)[source]¶
Bases:
EnergyTask- optional_params = ['software_kwargs', 'energy_kwargs', 'ignore_errors']¶
- required_params = ['software', 'label']¶
- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.MolecularIRC(*args, **kwargs)[source]¶
Bases:
FiretaskBase- optional_params = ['software', 'socket', 'software_kwargs', 'opt_kwargs', 'run_kwargs', 'constraints', 'ignore_errors', 'forward']¶
- required_params = ['xyz', 'label']¶
- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.MolecularOptimizationFailTask(*args, **kwargs)[source]¶
Bases:
OptimizationTask- optional_params = ['software_kwargs', 'opt_method', 'opt_kwargs', 'run_kwargs']¶
- required_params = ['software', 'label']¶
- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.MolecularOptimizationTask(*args, **kwargs)[source]¶
Bases:
OptimizationTask- optional_params = ['software_kwargs', 'opt_method', 'opt_kwargs', 'run_kwargs', 'constraints', 'sella', 'order', 'socket', 'time_limit_hrs', 'fmaxhard', 'ignore_errors', 'target_site_num', 'metal', 'facet']¶
- required_params = ['software', 'label']¶
- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.MolecularTSEstimate(*args, **kwargs)[source]¶
Bases:
FiretaskBase- optional_params = ['out_path', 'spawn_jobs', 'nprocs']¶
- required_params = ['rxn', 'ts_path', 'slab_path', 'adsorbates_path', 'rxns_file', 'repeats', 'path', 'metal', 'facet', 'name_to_adjlist_dict', 'gratom_to_molecule_atom_maps', 'gratom_to_molecule_surface_atom_maps', 'opt_obj_dict', 'vib_obj_dict', 'IRC_obj_dict', 'nslab', 'Eharmtol', 'Eharmfiltertol', 'Ntsmin', 'max_num_hfsp_opts']¶
- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.MolecularTSNudge(*args, **kwargs)[source]¶
Bases:
FiretaskBase- optional_params = ['forward_path', 'reverse_path', 'spawn_jobs', 'software', 'opt_method', 'sella', 'socket', 'software_kwargs', 'opt_kwargs', 'run_kwargs', 'constraints', 'ignore_errors']¶
- required_params = ['vib_traj', 'label']¶
- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.MolecularTSxTBOpt(*args, **kwargs)[source]¶
Bases:
OptimizationTask- optional_params = ['label', 'ignore_errors']¶
- required_params = ['xyz', 'slab_path', 'bonds', 'repeats', 'av_dists_tuple']¶
- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.MolecularVibrationsTask(*args, **kwargs)[source]¶
Bases:
VibrationTask- optional_params = ['software_kwargs', 'constraints', 'ignore_errors', 'socket']¶
- required_params = ['software', 'label']¶
- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- class pynta.tasks.OptimizationTask(*args, **kwargs)[source]¶
Bases:
FiretaskBase- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- pynta.tasks.TSnudge_firework(xyz, label, forward_path=None, reverse_path=None, spawn_jobs=False, software=None, opt_method=None, sella=False, socket=False, software_kwargs={}, opt_kwargs={}, run_kwargs={}, constraints=[], parents=[], out_path=None, ignore_errors=False)[source]¶
xyz is really the vibrational output data file out_path is a dud variable here
- pynta.tasks.TSxTBOpt_firework(xyz, slab_path, bonds, repeats, av_dists_tuple, out_path=None, label='', parents=[], ignore_errors=False)[source]¶
- class pynta.tasks.VibrationTask(*args, **kwargs)[source]¶
Bases:
FiretaskBase- run_task(fw_spec)[source]¶
This method gets called when the Firetask is run. It can take in a Firework spec, perform some task using that data, and then return an output in the form of a FWAction.
- Args:
- fw_spec (dict): A Firework spec. This comes from the master spec.
In addition, this spec contains a special “_fw_env” key that contains the env settings of the FWorker calling this method. This provides for abstracting out certain commands or settings. For example, “foo” may be named “foo1” in resource 1 and “foo2” in resource 2. The FWorker env can specify { “foo”: “foo1”}, which maps an abstract variable “foo” to the relevant “foo1” or “foo2”. You can then write a task that uses fw_spec[“_fw_env”][“foo”] that will work across all these multiple resources.
- Returns:
(FWAction)
- pynta.tasks.collect_firework(xyzs, check_symm, fw_generators, fw_generator_dicts, out_names, future_check_symms, parents=[], label='')[source]¶
- pynta.tasks.construct_constraint(d)[source]¶
construct a constrain from a dictionary that is the input to the constraint constructor plus an additional “type” key that indices the name of the constraint returns the constraint
- pynta.tasks.debug_fizzled(fw, trace, task)[source]¶
generate a new firework to replace the fizzled firework
- pynta.tasks.energy_firework(xyz, software, label, software_kwargs={}, parents=[], out_path=None, ignore_errors=False)[source]¶
- pynta.tasks.get_fw_traceback_task(fizzfw)[source]¶
get the traceback and task errored on for the fizzled firework
- pynta.tasks.optimize_firework(xyz, software, label, opt_method=None, sella=None, socket=False, order=0, software_kwargs={}, opt_kwargs={}, run_kwargs={}, constraints=[], parents=[], out_path=None, time_limit_hrs=inf, fmaxhard=0.0, ignore_errors=False, target_site_num=None, metal=None, facet=None, priority=1)[source]¶
- pynta.tasks.reconstruct_firework(new_task, old_task, task_list, full=True)[source]¶
reconstruct a firework replacing an old_task with new_task based on the task list full indicates whether all of the tasks should be included or just those starting from the replaced task
- pynta.tasks.reconstruct_task(task_dict, orig_task=None)[source]¶
regenerate a task based on the task_dict
- pynta.tasks.restart_opt_firework(task, task_list)[source]¶
generate a firework to restart an optimization firework from the trajectory file based on the optimization task and the full task list
- pynta.tasks.restart_wf(lpad, queue)[source]¶
lpad is a LaunchPad object queue is a boolean indicating if running in a queue or not
- pynta.tasks.vibrations_firework(xyz, software, label, software_kwargs={}, parents=[], out_path=None, constraints=[], socket=False, ignore_errors=False)[source]¶
- pynta.utils.filter_nonunique_TS_guess_indices(geoms, Es)[source]¶
Check for the symmetry equivalent structures in the given files
- Parameters:
geoms (list of paths to .xyz or .traj files to compare) –
- pynta.utils.get_unique_sym(geoms)[source]¶
Check for the symmetry equivalent structures in the given files
- Parameters:
geoms (list of paths to .xyz or .traj files to compare) –
- Returns:
idx_list – a list with prefixes of all symmetrically distinct sites
- Return type:
list(str)
- pynta.utils.get_unique_sym_indices(geoms)[source]¶
Check for the symmetry equivalent structures in the given files
- Parameters:
geoms (list of paths to .xyz or .traj files to compare) –
- pynta.utils.get_unique_sym_struct_index_clusters(geoms)[source]¶
Check for the symmetry equivalent structures in the given files
- Parameters:
geoms (list of Atoms objects to compare) –
- pynta.utils.get_unique_sym_struct_indices(geoms)[source]¶
Check for the symmetry equivalent structures in the given files
- Parameters:
geoms (list of Atoms objects to compare) –
- pynta.utils.get_unique_sym_structs(geoms)[source]¶
Check for the symmetry equivalent structures in the given files
- Parameters:
geoms (list of Atoms objects to compare) –
- pynta.utils.name_to_ase_software(software_name)[source]¶
go from software_name to the associated ASE calculator constructor
- pynta.transitionstate.determine_TS_construction(reactant_names, reactant_mols, product_names, product_mols)[source]¶
determine the direction to generate the TS guess in and the order in which the reactants will be placed on the surface returns forward a boolean indicating if the reaction should be estimated in the forward (True) or reverse (False) direction and ordered_reacting_species which is a list of species in the order they should be placed on the surface
- pynta.transitionstate.estimate_deq_k(labels, dwell, forward_template, reverse_template, template_name, template_reversed, broken_bonds, formed_bonds, sitetype=None)[source]¶
Estimate the equilibrium bond length and force constant for broken/forming bonds 0–(1–2)–3
- pynta.transitionstate.estimate_deq_k_fixed_surf_bond(labels, dwell, forward_template, reverse_template, template_name, template_reversed, broken_bonds, formed_bonds, sitetype=None)[source]¶
Estimate the equilibrium bond length and force constant for surface bonds
- pynta.transitionstate.generate_constraints_harmonic_parameters(tsstructs, adsorbates, slab, forward_template, reverse_template, template_name, template_reversed, ordered_names, reverse_names, mol_dict, gratom_to_molecule_atom_maps, gratom_to_molecule_surface_atom_maps, nslab, facet, metal, cas)[source]¶
Generate constraints and harmonic parameters for the harmonically forced optimization
- pynta.transitionstate.get_surface_forming_bond_pairings(tsstructs, atom_bond_potential_lists, site_bond_potential_lists, constraints_list, site_bond_dict_list, cas)[source]¶
Identify unique site targets for the harmonically forced optimization
- pynta.transitionstate.get_unique_TS_structs(adsorbates, species_names, cas, nslab, num_surf_sites, mol_dict, gratom_to_molecule_atom_maps, gratom_to_molecule_surface_atom_maps, facet, metal, gas_height=5.0)[source]¶
Generate unique initial structures for TS guess generation
- pynta.transitionstate.get_unique_optimized_adsorbates(rxn, adsorbates_path, mol_dict, cas, gratom_to_molecule_surface_atom_maps, nslab)[source]¶
load the adsorbates associated with the reaction and find the unique optimized adsorbate structures for each species returns a dictionary mapping each adsorbate name to a list of ase.Atoms objects
Indices and tables¶
Acknowledgments¶
If you are using pynta or you wish to use it, let me know!
Copyright Notice¶
For five (5) years from 1/19/2021 the United States Government is granted for itself and others acting on its behalf a paid-up, nonexclusive, irrevocable worldwide license in this data to reproduce, prepare derivative works, and perform publicly and display publicly, by or on behalf of the Government. There is provision for the possible extension of the term of this license. Subsequent to that period or any extension granted, the United States Government is granted for itself and others acting on its behalf a paid-up, nonexclusive, irrevocable worldwide license in this data to reproduce, prepare derivative works, distribute copies to the public, perform publicly and display publicly, and to permit others to do so. The specific term of the license can be identified by inquiry made to National Technology and Engineering Solutions of Sandia, LLC or DOE.
NEITHER THE UNITED STATES GOVERNMENT, NOR THE UNITED STATES DEPARTMENT OF ENERGY, NOR NATIONAL TECHNOLOGY AND ENGINEERING SOLUTIONS OF SANDIA, LLC, NOR ANY OF THEIR EMPLOYEES, MAKES ANY WARRANTY, EXPRESS OR IMPLIED, OR ASSUMES ANY LEGAL RESPONSIBILITY FOR THE ACCURACY, COMPLETENESS, OR USEFULNESS OF ANY INFORMATION, APPARATUS, PRODUCT, OR PROCESS DISCLOSED, OR REPRESENTS THAT ITS USE WOULD NOT INFRINGE PRIVATELY OWNED RIGHTS.
Any licensee of this software has the obligation and responsibility to abide by the applicable export control laws, regulations, and general prohibitions relating to the export of technical data. Failure to obtain an export control license or other authority from the Government may result in criminal liability under U.S. laws.
License Notice¶
Copyright 2021 National Technology & Engineering Solutions of Sandia, LLC (NTESS). Under the terms of Contract DE-NA0003525 with NTESS, the U.S. Government retains certain rights in this software.